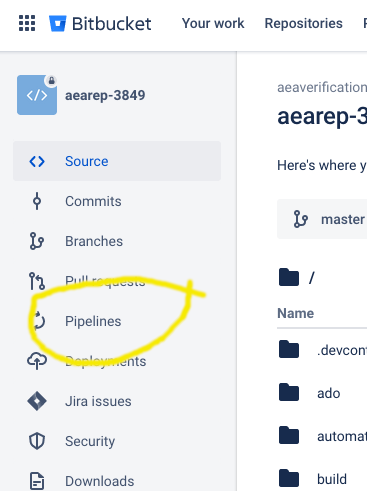
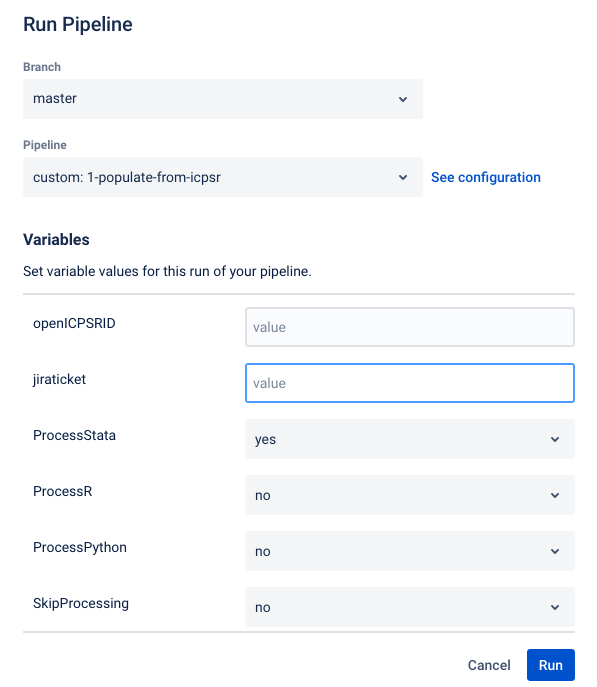
Creating a repository and running pipeline#
Warning
In some instances, somebody else has already created a repository. Always check first if the Bitbucket short name is already filled out. If it is, do not run the pipeline scripts again. This can overwrite or destroy changes already made by someone working on the case.
If the Bitbucket short name is filled out, skip this section and go to Collecting information!
Creating a repository#
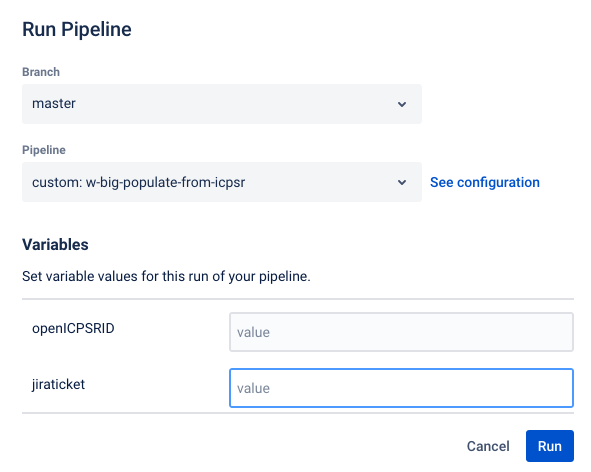
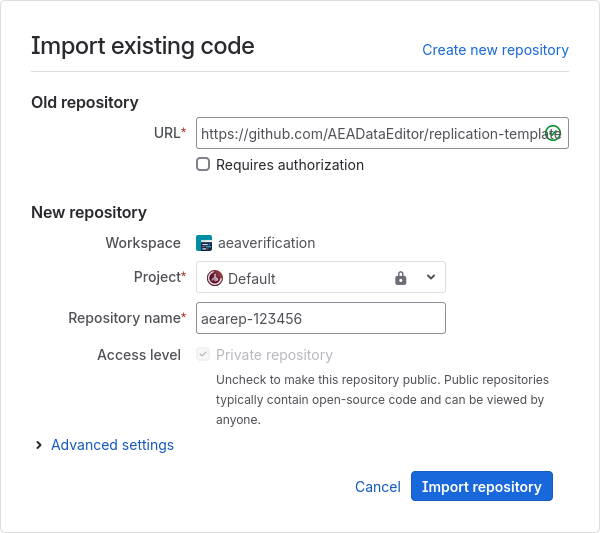
start by creating a repository using the import method
copy-paste from this URL to the URL field (this is also available in the Jira dropdown “Shortcuts”)
https://github.com/AEADataEditor/replication-template
the repository name should be the name of the JIRA issue, in lower case (e.g.,
aearep-123)Be sure that
aeaverificationis always the “owner” of the report on Bitbucket.The Project should be the
DEFAULTproject, unless you are explicitly asked to create it in a different one (currently only “JEP” and “JEL”)Keep the other settings (in particular, keep this a private repository).
Click
Import RepositoryKeep this tab open!
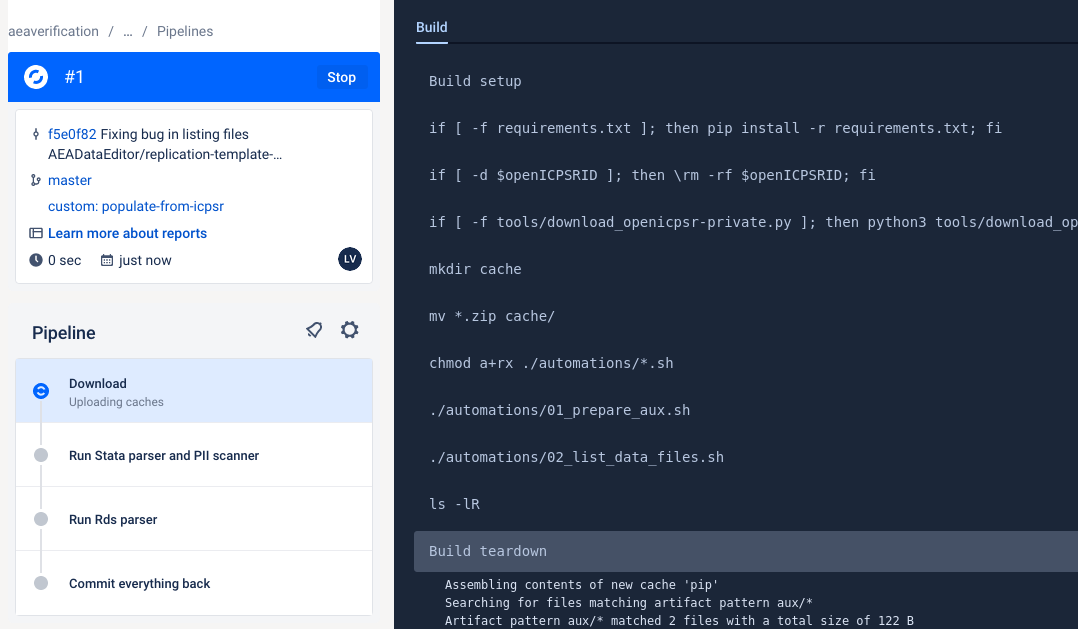
We have now created a Bitbucket repo named something like
aearep-123that has been populated with the latest version of the LDI replication template documents!
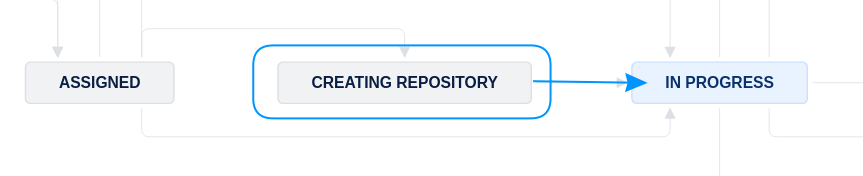
Next step#
Once everything has completed to your satisfaction,
fill out the
Bitbucket short namefield (e.g.,aearep-1234)Move the Jira issue forward to
In Progress.