Prepare the working area#
Note
Link to JIRA: https://aeadataeditors.atlassian.net/jira (requires login).
Computer access: Access computers appendix.
Before we can verify code, data, and documentation, we need to get the code and data onto your “working area”.
Note
You now need to decide on what computer you are going to do the data analysis - that should be the place you do the next few steps. See Access Computer for details. This is because the git setup we use does not allow you to include the data files in the Bitbucket repository, so when you download the replication package from openICPSR or elsewhere, they do not get added to the Bitbucket repository.
Clone the Bitbucket repository onto the CCSS server you are working on
git clone https://yourname@bitbucket.org/aeaverification/aearep-xxx.git
Alternatively: If you have done the full Bash setup, you can run
aeagit xxxVerify that the code is present, i.e., that the automated scripts run during the Code Ingest worked. If they did not, you need to switch to the “Manual steps”!
From the JIRA issue, download and add Manuscript, Data and Code Availability Form (DCAF).
Download from Jira issue attachments. The manuscript is often called
PDF_Proof.pdf.
Add the manuscript, and any response by the authors (if a revision)
Add them to the Git repo
Be sure to
git pushit all to Bitbucket, with a meaningful commit message.git add PDF_Proof.pdf DataCodeAvailability.pdf git commit -m "added manuscript and DCAF" git push
All actions on BioHPC will be performed in a terminal. Depending on whether you connect with SSH, VNC, or Visual Studio Code, details may differ (see Access Computer for details). We suggest connecting via SSH or using Visual Studio Code (which uses SSH in the background), for simplicity. It is assumed that you have done Bash setup.
We suggest doing all first steps (adding Manuscript, etc.) on CCSS or your laptop.
If using SSH, you are already at the “terminal”. If using VNC, choose it from the application menu. If using [Visual Studio Code to connect to BioHPC], follow instructions from the Github Codespaces tab!
Change directory to the common workarea:
bash cd /home2/ecco_lv39/WorkspacePopulate BioHPC with the Bitbucket repo (yes, this is a bit weird)
Use the LDI short-cut command
aeagit 123 http
to clone the Bitbucket repository for
aearep-123onto BioHPC (this executedgit clone (URL)/aearep-123behind the scenes).If this is the first repository you run this, you may need to configure authentication. Follow the instructions from the
aeagitcommand.Once the
aeagit 123has completed, it show a bunch of error messages (maybe), and a commandcd aearep-123. Copy and paste that command, entering the directory where you just cloned the repo. All subsequent actions should be done in that window.
Verify that the code is present, i.e., that the automated scripts run during the Code Ingest worked. If the scripts did not work, run them now:
Download the project from openICPSR, using the script. See the details in the appendix.
The downloaded ZIP file (
123456.zip) should have been unzipped automatically (the terminal output will tell you).Then run the ingest scripts:
./tools/pipeline-steps1-4.sh 123456 git push
Add the manuscript, and any response by the authors (if a revision) - you should already have done this as per the CCSS tab!
All actions in Github Codespaces (CS for short) will be performed in Visual Studio Code (VSC): the left pane for file and Git actions, in the Terminal, or in the text editor. To connect to CS, see [instructions to come]. It is assumed that you will run this from your laptop, but it can be run from any internet-connected computer. The particular template you use for CS already has the Bash setup done for you.
Use the Terminal built into VSC (
Menu->Terminal->New Terminal). By default, the Terminal should run abashshell.Populate CS with the Bitbucket repo (yes, this is a bit weird)
Use the LDI short-cut command
aeagit 123
to clone the Bitbucket repository for
aearep-123onto CS (this executedgit clone (URL)/aearep-123behind the scenes).If this is the first repository you run on this CS instance, you may need to configure authentication. Follow the instructions from the
aeagitcommand.Once the
aeagit 123has completed, it will open a new VSC windows. All subsequent actions should be done in that window.
Verify that the code is present, i.e., that the automated scripts run during the Code Ingest worked. If the scripts did not work, run them now:
Download the project from openICPSR, using the script. See the details in the appendix.
The downloaded ZIP file (
123456.zip) should have been unzipped automatically (the terminal output will tell you).Then run the ingest scripts:
./tools/pipeline-steps1-4.sh 123456 git push
Add the manuscript, and any response by the authors (if a revision)
Copy the manuscript to the CS. There are two ways to do this:
Drag-and-drop the downloaded manuscript into the file pane of VSC.
Use the
ghcommand line tool from a non-VSC terminal (on your local computer):gh cs cp PDF_Proof.pdf remote:/workspaces/aearep-123
Add the manuscript to the Git repo
In the terminal:
git add PDF_Proof.pdf; git commit 'Manuscript'; git push
Or use the VSC Git tools
If the automated population of the author’s code directory did not work, you will need to manually download the replication package. Try to do this first using scripts.
Downloading using scripts
See the details in the appendix. You can do this on CCSS, BioHPC, or on Github CS.
Downloading using a browser
If you are still unsuccesful at this point, try this manual method (typically, on CCSS)
Download the code (and data) from openICPSR (typicaly for most cases). See details in the appendix for instructions on downloading these materials. Typically called
111234.zip.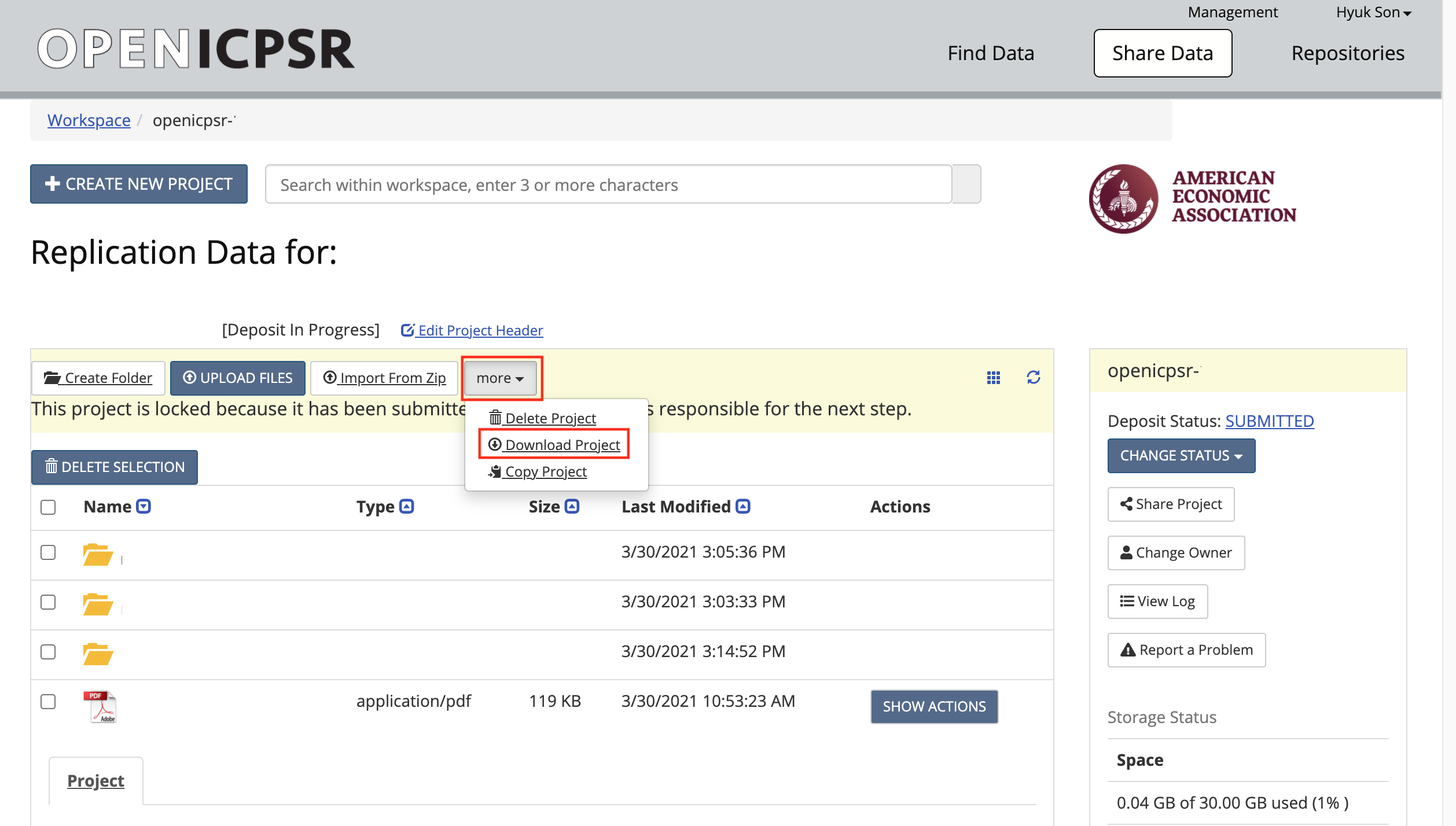
Copy/paste the downloaded openICPSR ZIP file into the local copy of the
aearep-123repositoryThe ZIP file should be called something like
111234.zip. Note: it might look like a folder, but it is not! (on Windows)The ZIP file will be wherever your browser downloads materials - probably your
Downloadfolder.
Next manual steps
The local repository should now have the relevant LDI replication template materials and the openICPSR ZIP file containing the replication materials provided by the authors.
If uploading to CS, copy the downloaded ZIP file into the CS. There are two ways to do this:
Drag-and-drop the downloaded openICPSR ZIP file into the file pane of VSC.
Use the
ghcommand line tool from a non-VSC terminal (on your local computer):gh cs cp 111234.zip remote:/workspaces/aearep-123
(adjust accordingly)
Unzip the openICPSR folder under a folder named for the openICPSR repostory number.
From bash:
unzip -n 111234.zip -d 111234
On Windows, right-click and select “Extract all”. When asked, do not overwrite files.
On OSX, double-click. When asked, do not overwrite files.
The individual files that are part of the replication package should now be in a subdirectory (e.g,
111234, the openICPSR repository number).Perform a
git add:git add 111234should do the right thing.
Add the manuscript, and any response by the authors (if a revision)
Add them to the Git repo
git add PDF_Proof.pdf DataCodeAvailability.pdf 111234 git commit -m "Adding manuscript, DCAF, and code" git push
Clean-up: Delete (
git rm) unused files from the template!Example:
git rm README.md template-config.Rif the replication archive does not contain any R files (you can do this at any time before writing the Preliminary Report)Make this a commit:
git commit -m "Deleting unnecessary files" git push
The root of the repository should contain only our files (i.e., REPLICATION.md, etc.), the manuscript files (main manuscript, any online appendices and README files provided through the JIRA ticket), and the code.
Example:
111234/ code-check.xlsx config.do PDF_Proof.PDF PII_stata_scan.do DataCodeAvailability.pdf REPLICATION.md
Be sure to
git pushit all to Bitbucket.
Note
git push
Check with
git statusthat there are no uncommited files.