Reproducibility with confidential data: The experience of BPLIM
Background
The Banco de Portugal Microdata Laboratory (BPLIM) was established in 2016 with the primary goal of promoting external research on the Portuguese economy by making available data sets collected and maintained by Banco de Portugal (BdP). By making this information available to researchers from around the world, BdP aims to support the development of evidence-based policies and insights that can benefit the Portuguese economy and society. However, given that some of these data sets contain highly sensitive information BPLIM had to implement a data access solution that preserved the confidentiality of the data.
The common approach by other Research Data Centers that make confidential data available for research involves the provision of on-site access to accredited external researchers in a secure computing environment. However, in the case of BPLIM this approach was deemed undesirable for two reasons. First, because it would limit access to a handful of researchers who were able to come to the Bank's premises. Second, because there were still concerns that a breach of confidentiality might occur if individuals from outside the bank could gain access to original data sets that contained confidential information.
After an internal debate at the Bank it was decided that the solution to be adopted by BPLIM had to be based on the following principles:
- access free of charge and only for scientific purposes;
- all data should be analyzed on the servers of the Bank;
- external researchers were granted remote access to the server;
- confidential datasets placed on the server had to always be perturbed/masked;
- researchers could always ask BPLIM staff to run their scripts on the original data.
The general workflow defined for data access at BPLIM was the following. After a research project is approved and the external researchers are accredited an account is opened on the BPLIM external server. External researchers gain access to a computing environment that does not allow users to transfer files to and from the server. They have access to a restricted area where standard software such as Stata, R, Julia and Python are available. Since there is no connection to the internet, installation of specific packages has to be requested from the staff. The datasets for the project are placed in a read-only folder. For the confidential datasets what is placed in the account of the researcher are perturbed versions of the data (noise is added to the original data). The researcher implements all scripts based on the data he/she has available and produces the (non-valid) outputs required for the project. Once researchers complete this task, they can ask BPLIM staff to rerun their scripts, this time using the original confidential data. For this process to be successful BPLIM staff must first run the scripts using the same data as the researcher to verify that the scripts written by the researchers reproduce exactly the outputs (typically graphs and/or tables). This process is done in a different server (BPLIM internal server). Only upon completion of this first step can BPLIM staff modify the scripts, this time to read the original data and regenerate the intended outputs . These outputs are then subject to standard output control checks for confidential data and delivered to the researcher.
Main Thoughts
Over time we have come to realize that this somewhat cumbersome process of running the code thrice, first by the researcher on the perturbed data, second by BPLIM staff again on the perturbed data, and finally, on the original data was in fact an exercise on reproducibility. Even though the reproducibility check was on the perturbed data it was already a very good assurance that a reproducibility check would hold on the original data.
We realized that a great deal of our work involved reproducing the results obtained by the researchers with the perturbed data and that led us to look at ways to improve our workflow. It became obvious that the process could be streamlined and would be more efficient if researchers adhered to the best practices on reproducibility. Hence, as part of our strategy, we have decided to raise awareness of our researchers to the need of implementing good practices on reproducible research. We have been doing this by several means. For example, we have held practical workshops which are designed to enhance the skills of our researchers. For these workshops we invite leading experts to present best practices and recent developments on data analysis. We also provide direct advice to the researchers, prepare templates and documentation, and make available tools that facilitate the analysis of our datasets (particularly for more complex tasks such as building a panel or calculation of specific variables).
On the other end, it was also obvious that there was margin for improvement on our work sequence. One possible improvement was the assurance that the computing environment used by the researcher on BPLIM's external server was identical to that used by BPLIM staff when reproducing the code. Thus, for the case of researchers that work with open-source software, we have been incentivizing researchers to work with Singularity containers. This facilitates our work because we are sure that our reproducibility check is implemented in the same self-contained environment that was used by the researcher. Researchers that use Stata can resort to containers but in that case, it is easier to control the environment because we install all packages on a folder that is specific to each project and have developed tools that facilitate comparison of the Stata ado files across environments. (all tools are publicly available and can be found at https://github.com/BPLIM/Tools/tree/master/ados/General.)
More recently, we have worked on shifting the burden of the reproducibility check to the researcher itself. We are developing an application that we are presently testing with a select number of researchers. The application is targeted mainly at researchers that use BPLIM's (perturbed) confidential datasets but we hope to eventually convince all other users to take advantage of it. To illustrate, we provide a screenshot of the application:
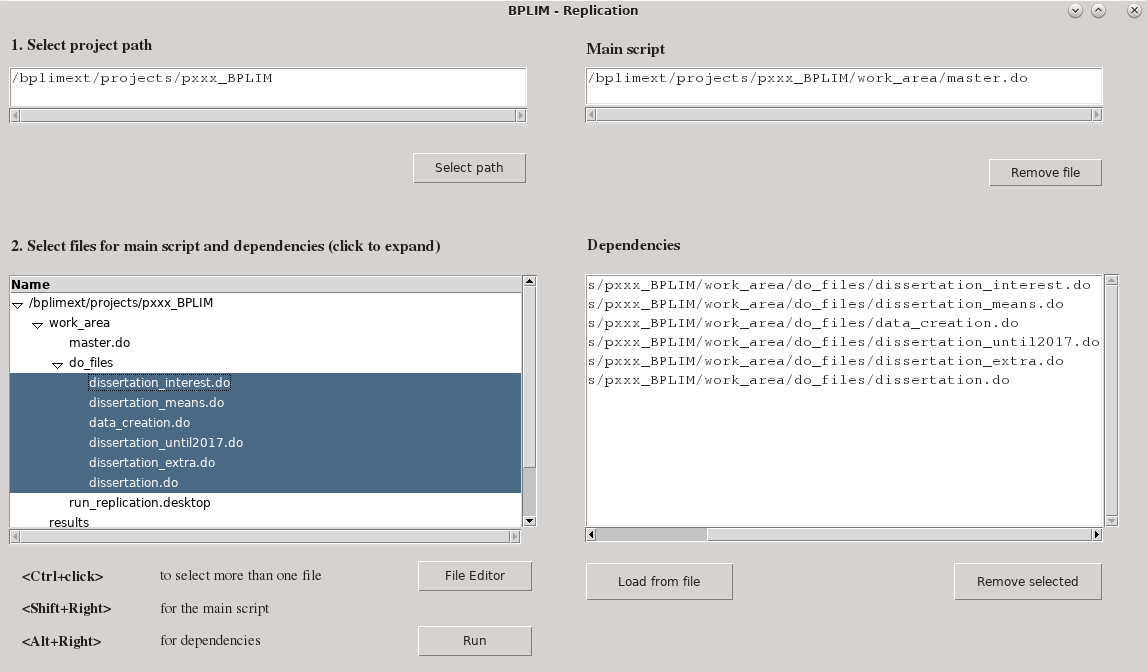
Figure - BPLIM Replication tool – selecting input files
Before requesting a replication from BPLIM using the original data the researcher must first validate his/her code by successfully submitting the scripts through BPLIM's Replication application. The process involves selecting the main script as well as all the required dependencies created by the researcher. The folder structure used by the researcher is replicated and the BPLIM datasets have to be read from the (read-only) data folder. All intermediary output files must be created during the replication (it is however possible to start from an intermediary output file. In that case, the intermediary file must have been validated in a prior run. BPLIM will then copy the file to the (read-only) data folder.).
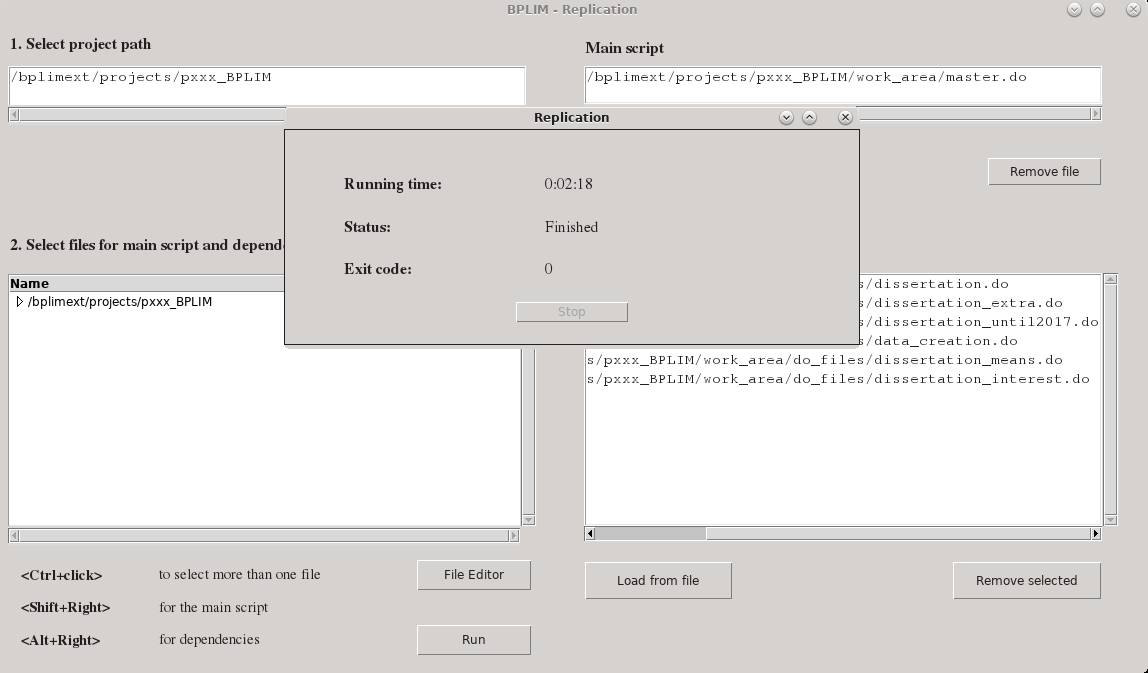
Figure - The BPLIM Replication tool - finished task
The researcher then uses the application to run the code (see Figure 2 for a completed run). The code must run from top-to-bottom and produce no errors. If the run is successful, then implementation on the original data requires only that BPLIM staff changes the relative paths to the data folder and rerun the code. A side advantage of this process is that it automatically produces a replication package for the researcher. Stored in the folder are all replication scripts, the output files, as well as two additional files, one fully characterizing the software environment, and another json file containing a listing of all scripts used in the replication. If we add the definition file used to produce the container (or a listing of all packages and respective versions) then we have a full replication package (except for the data).
Conclusion
Our goal at BPLIM is to make sure that all researchers create their replication packages as an integrated part of their research process. The fact that we are the ones running the code on the original data should be seen as an opportunity to request that researchers make reproducible code while implementing their research.
In the ideal situation that we envision, researchers download a template for the definition file of the Singularity container, customize that template by adding and testing the packages they need, and share with us the definition file. Based on that definition file we build the container for the project and make it available on our external server. The researcher then uses the container to implement the analysis and when he/she is ready to obtain results based on the original data he must first validate the scripts using our application. The researcher can go through this process multiple times and each time a replication package will be created. Apart from the data, all files can be publicly shared, and the replication package created at BPLIM should be easily customized for submission to any data editor.
We are already implementing this solution for all new projects that use confidential data. However, we hope that over time we can convince all researchers at BPLIM to work with Singularity containers and go through the same validation steps that are needed for projects that deal with confidential data.
Projects that are implemented in BPLIM have additional advantages when it comes to reproducibility. First, because all BPLIM datasets are versioned and registered with a Digital Object Identifier (DOI) we are sure that the original data is exactly identified. Second, the computing environment is stable, and the software packages used by researchers are specific to each project. Finally, if external researchers have used BPLIM confidential data then there is an assurance that their code was reproduced at some point.
Ultimately, whether the scientific work is reproducible depends on the researchers. But we hope that integrating reproducibility into the research process with confidential data, provides a way to alleviate the inconvenience of third parties that cannot access the original data and want to verify reproducibility of the results.